#bioinformatics tools
Explore tagged Tumblr posts
Text
#bioinformatics tools#application of bioinformatics#bioinformatics data#bioinformatics database#next generation sequencing data analysis#genomic data analysis#bioinformatics analysis
0 notes
Text
Maximizing Efficiency: Best Practices for Using Sequencing Consumables

By implementing these best practices, researchers can streamline sequencing workflows, increase throughput, and achieve more consistent and reproducible results in genetic research. Sequencing Consumables play a crucial role in genetic research, facilitating the preparation, sequencing, and analysis of DNA samples. To achieve optimal results and maximize efficiency in sequencing workflows, it's essential to implement best practices for using these consumables effectively.
Proper planning and organization are essential for maximizing efficiency when using Sequencing Consumables. Before starting a sequencing experiment, take the time to carefully plan out the workflow, including sample preparation, library construction, sequencing runs, and data analysis. Ensure that all necessary consumables, reagents, and equipment are readily available and properly labeled to minimize disruptions and delays during the experiment.
Optimizing sample preparation workflows is critical for maximizing efficiency in sequencing experiments. When working with Sequencing Consumables for sample preparation, follow manufacturer protocols and recommendations closely to ensure consistent and reproducible results. Use high-quality consumables and reagents, and perform regular quality control checks to monitor the performance of the workflow and identify any potential issues early on.
Utilizing automation technologies can significantly increase efficiency when working with Sequencing Consumables. Automated sample preparation systems and liquid handling robots can streamline repetitive tasks, reduce human error, and increase throughput. By automating sample processing and library construction workflows, researchers can save time and resources while improving consistency and reproducibility in sequencing experiments.
Get More Insights On This Topic: Sequencing Consumables
#Sequencing Consumables#DNA Sequencing#Laboratory Supplies#Genetic Analysis#Next-Generation Sequencing#Molecular Biology#Research Tools#Bioinformatics
2 notes
·
View notes
Text
ChatGPT for Academia: Anatomy of Advanced ChatGPT Prompt


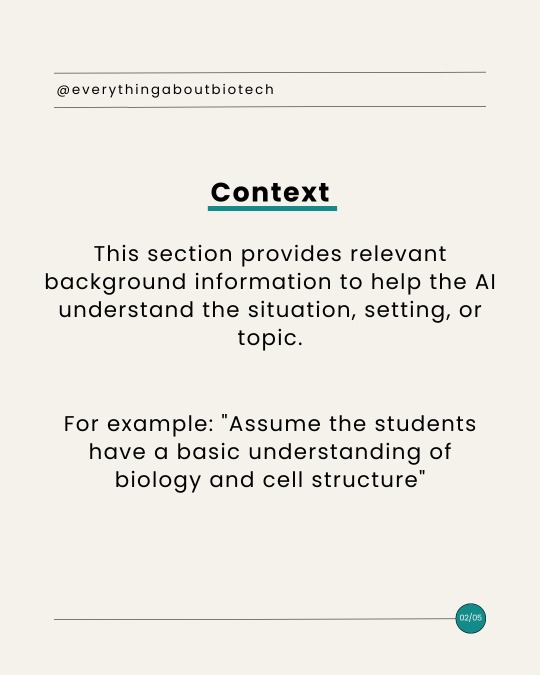


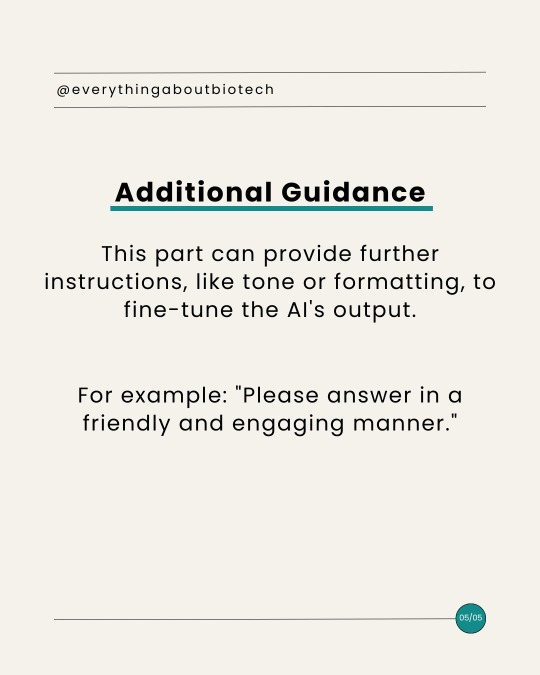


#academic chat#chatgpt#academic chatter#chatbot#ai tools#ai#biotechnology#science#bioinformatics#research#tips#communication#writing#skills#academia#design#bard#aiforeducation#academic#academic writing
8 notes
·
View notes
Text
How to Write a Python Script
Python is a high-level programming language that many organizations use. Developers can use it to build sites, analyze data, perform tasks and more. A Python script is a set of commands written in a file that runs similarly to a program, allowing the file to perform a specific function.
The best and easiest way to write code is to use data apps for Python scripts. The right app can save you hours while reducing the risk of error. But even if you use data apps for Python scripts, it pays to know how to write them from scratch.
In this blog, you'll learn how to write a simple Python Script with one of the most famous coding exercises in the world.
Organize Your Scripts
The first step is to create a folder for your Python scripts. Naming conventions are flexible. The key is to choose a folder hierarchy that makes sense to you. For simplicity, consider making a separate folder on your desktop. You can name it "python_workshop."
Creating Your First Script
To start your script, open up Notepad++ and create a new file. Then, click File>Save As.
Save your script in your newly created "python_workshop" folder and name it "exercise_hello_world.py." Pay attention to that file extension!
Write Your Code
On the first line, type out:
# Author: YOUR NAME and email address
Fill out the code with the relevant information to make the script run properly.
On the next line, type:
# This is a script to test that Python is working.
Add an empty line to create your next line of code. It should read:
print("Hello world from YOUR NAME").
Save your file.
Running the Script
Open a new Terminal window. You should see:
C:\Users\YOURNAME>
After you see your name, type in "CD desktop." Hit "Enter," and you can type in the name of your "python_workshop" folder. Hit "Enter" again, and type in the name of the script file you just created.
After hitting "Enter," you should see that little "hello world" message you created in your script.
Read a similar article about chatgpt and spreadsheets here at this page.
#low code sql tool#data apps for python scripts#python integration for enterprise#python for bioinformatics
0 notes
Text
I feel like I’m missing something here bc there should be a way to easily compare species from two different BLAST searches, right????? All I’m trying to do is get a list of species that have a hit for a homologue of one or both of two different proteins and see which ones have what
but the only way I can find how to do this is to download both BLAST searches and write a script to cross reference the species info, which I can do, but I feel like there should be a shortcut?????????
#idk how to tag this#might have to ask on quora ugh#biology#biology question#bioinformatics#456 words#also doesn’t technically have to be blast results it can be a homology search on uniprot or something#but yeah this seems like something that would be done regularly enough to merit a shortcut or tool
0 notes
Text

[The Idiot's Guide to Effective Population Size
This is a reference manual for the elegant, yet hideously complex concept of effective population size (Ne), inspired by a classic, self-published manual of automotive repair ‘for the compleat idiot’. The Guide is timely, given the recent Kunming-Montreal Global Biodiversity Framework, where 196 Parties committed to tracking genetic diversity—and estimating Ne—for all species. Ne is a human construct, but a useful one that allows us to capture diverse aspects of an organism's biology in a single number. The Guide collates in one location factual information about effective population size, with a focus on topics of practical relevance to scientists and managers studying real populations; it covers definition, computation and estimation of effective size, both demographically and genetically. As appropriate, the reader is directed to other primary sources for more details. A ‘Don't Do These Things’ section lists several ill-advised approaches to dealing with Ne, and an Appendix provides useful tools and practical suggestions for interested users. A special section considers both possibilities and challenges presented by the genomics revolution. Availability of vast numbers of genetic markers increases precision, but less than some might think, and simultaneously introduces new challenges involving filtering and bioinformatics processing. As annotated genomes become more common for non-model species, opportunities are opened to address qualitatively different questions, including reconstructing historical changes in Ne through time.]
Waples (2025)
250 notes
·
View notes
Text
@hellsite-proteins
“Funny shapes”
“The analyses of each protein are hilarious even if you don’t know anything about proteins”
“AlphaFold is an incredibly powerful tool in bioinformatics and is a contender for the Nobel Prize in upcoming years, but is underappreciated by the general public. Seeing it put to use in such a silly and fun way is both refreshing and educational!”
“Turns posts into big squiggly lines :3”
@aroace-everyday
(no propaganda submitted)
178 notes
·
View notes
Text
My last project for this semester is for a multimedia databases class and it's a simple "Pick three recent papers that are relevant to this class and have a shared topic and write a summary of them". Since I'm in bioinformatics, I decided to use this as an opportunity to read up on protein folding methods, and I have gotten bizarrely obsessive about explaining everything I can about the papers I chose.
I wrote a whole page just going over the Critical Assessment of Structure Prediction (CASP) that's used to measure the accuracy of protein folding tools, and set aside a whole section in the first paper summary where I gave a brief explanation about certain types of protein folds.
I'm making this post because I realized two of the papers I'm using are from the same author and I just emailed them to ask for a comment on why one of the tools they made is no longer available. I'm assuming it's because the later tool they made makes the older one obsolete, but I want to get confirmation before I say anything.
10 notes
·
View notes
Note
Hi there! I hope this doesn’t sound antagonistic (I’m not trying to be, just playing the devils advocate. Also, my background is more in bioinformatics, so I’m not sure if that counts as “layman” or not, but I do handle a lot of data and databases) but a lot of blowback regarding AI stuff is the sheer desperation a lot of artist feel regarding the scraping of art. Work really has dried up for many artists directly as a result from AI, and it doesn’t help a lot of studios have been laying off people. The people at the working at Nightshade and Glaze understand it’s not a perfect tool or solution, but, like they clarified before, it does provide some friction and it’s still being worked on. Personally, I would also like some of the energy directed to making some legislation happened, but it’s going to take time and a concerted movement before that happens.
May I ask what are your thoughts on this? I would love to hear more from you! I think most of the time people act rashly is from desperation more than stupidity.
any "legislation" around AI art would take the form of corporations using this as an opportunity to expand IP and copyright law in ways that would only benefit them (corporations) and directly harm everyone else, including independent artists
the Luddites were wrong
57 notes
·
View notes
Text
In the Biopunk Future AIs are extensively used to process the extensive amounts of genetic data, from raw genome sequences to simulations of proteins and metabolism beyond what is available today (real life note; in my workplace, we have piles of hard drives full of genetic data). There could be no accessible genetic engineering without massive leaps in bioinformatics.
These AIs work mostly in the background, without much human (or non-human) input beyond what they're asked to do. They try to be as accurate as possible while solving complex problems such as balancing neuron growth and the tempo for industrial biochemical production in fruit trees. But when working with genomes of billions of base pairs and countless intricate dances of proteomics and metabolomics, inevitably, some hallucinations will occur even in the most perfectly edited genomes. Just a couple base pairs out of place, just a protein that is folded a little funky.
This happens most often with hobbyist, that is, biopunk tools, which often don't have the strict guardrails and raw power of corporate or state biotech. Biopunks welcome these small ghosts in the genome, they love them and appreciate when they're found and they're harmless. It makes a genome that has been otherwise completely created and edited in a silicon machine to feel alive.
#cosas mias#soft biopunk#biopunk#I have a vague idea of introducing a sentient biological machine as a character but I might leave it in the background
8 notes
·
View notes
Text
Free online courses for bioinformatics beginners
🔬 Free Online Courses for Bioinformatics Beginners 🚀
Are you interested in bioinformatics but don’t know where to start? Whether you're from a biotechnology, biology, or computer science background, learning bioinformatics can open doors to exciting opportunities in genomics, drug discovery, and data science. And the best part? You can start for free!
Here’s a list of the best free online bioinformatics courses to kickstart your journey.
📌 1. Introduction to Bioinformatics – Coursera (University of Toronto)
📍 Platform: Coursera 🖥️ What You’ll Learn:
Basic biological data analysis
Algorithms used in genomics
Hands-on exercises with biological datasets
🎓 Why Take It? Ideal for beginners with a biology background looking to explore computational approaches.
📌 2. Bioinformatics for Beginners – Udemy (Free Course)
📍 Platform: Udemy 🖥️ What You’ll Learn:
Introduction to sequence analysis
Using BLAST for genomic comparisons
Basics of Python for bioinformatics
🎓 Why Take It? Short, beginner-friendly course with practical applications.
📌 3. EMBL-EBI Bioinformatics Training
📍 Platform: EMBL-EBI 🖥️ What You’ll Learn:
Genomic data handling
Transcriptomics and proteomics
Data visualization tools
🎓 Why Take It? High-quality training from one of the most reputable bioinformatics institutes in Europe.
📌 4. Introduction to Computational Biology – MIT OpenCourseWare
📍 Platform: MIT OCW 🖥️ What You’ll Learn:
Algorithms for DNA sequencing
Structural bioinformatics
Systems biology
🎓 Why Take It? A solid foundation for students interested in research-level computational biology.
📌 5. Bioinformatics Specialization – Coursera (UC San Diego)
📍 Platform: Coursera 🖥️ What You’ll Learn:
How bioinformatics algorithms work
Hands-on exercises in Python and Biopython
Real-world applications in genomics
🎓 Why Take It? A deep dive into computational tools, ideal for those wanting an in-depth understanding.
📌 6. Genomic Data Science – Harvard Online (edX) 🖥️ What You’ll Learn:
RNA sequencing and genome assembly
Data handling using R
Machine learning applications in genomics
🎓 Why Take It? Best for those interested in AI & big data applications in genomics.
📌 7. Bioinformatics Courses on BioPractify (100% Free)
📍 Platform: BioPractify 🖥️ What You’ll Learn:
Hands-on experience with real datasets
Python & R for bioinformatics
Molecular docking and drug discovery techniques
🎓 Why Take It? Learn from domain experts with real-world projects to enhance your skills.
🚀 Final Thoughts: Start Learning Today!
Bioinformatics is a game-changer in modern research and healthcare. Whether you're a biology student looking to upskill or a tech enthusiast diving into genomics, these free courses will give you a strong start.
📢 Which course are you excited to take? Let me know in the comments! 👇💬
#Bioinformatics#FreeCourses#Genomics#BiotechCareers#DataScience#ComputationalBiology#BioinformaticsTraining#MachineLearning#GenomeSequencing#BioinformaticsForBeginners#STEMEducation#OpenScience#LearningResources#PythonForBiologists#MolecularBiology
9 notes
·
View notes
Text
Reference saved in our archive
An interesting potential diagnostic tool that deserves further study. Could be a clue to diagnosing or preventing long covid with some further understanding.
Abstract Objective As one of the remarkable host responses to SARS-CoV-2 infection, circulating microRNAs (miRNAs) represent important diagnostic and prognostic diseases biomarkers. The study is a step towards highlighting the role of miRNAs in COVID-19 pathogenesis and severity.
Methods In this case-control study, miRCURY LNA miRNA PCR plasma panel (168 miRNAs) was applied and the expression of the altered miRNAs was then analysed by quantitative real time PCR for 120 COVID-19 patients (30 mild, 30 moderate, 30 severe, and 30 critical) and 30 healthy subjects.
Results The initial screening showed that 30 miRNAs displayed altered expression, out of them, only eleven miRNAs (miR-885-5p, miR-141-3p, miR-21-5p, miR-127-3p, miR-99b-5p, let-7d-3p, miR-375, miR-1260a, miR-139-5p, miR-28-5p and miR-34a-5p) were dysregulated in the plasma of COVID-19 patients; all of them were significantly overexpressed. By applying ROC curve analysis, AUC for the eleven miRNAs were ranged from 0.65 to 0.83, and the AUC for the combined miRNAs was 0.93. Ten miRNAs (miR-141-3p, miR-181a-5p, miR-221-3p, miR-223-5p, miR99b-5p, Let-7d-3p, miR-375, miR-199a-5p, miR-139-5p and miR-28-5p) exhibited a significant change in their expression between different severity groups. Patients with positive outcome were found to have increased miR-375 and decreased miR-99b-5p expression levels. Bioinformatic prediction showed that, out of the eleven dysregulated miRNAs, five miRNAs (miR-139-5p, −34a-5p, −28-5p, −21-5p and −885-5p) have the ability to regulate at least two genes related to COVID-19 according to KEGG database.
Conclusion miRNAs are dysregulated in COVID-19 patients and associated with severity degree and patients’ outcome.
#mask up#public health#wear a mask#pandemic#wear a respirator#covid#still coviding#coronavirus#sars cov 2#covid 19
7 notes
·
View notes
Note
Hey Meg! I was just catching up on your posts and wow, congrats on publishing the paper! I'm so excited for you and how far you'll go (it's so cool that you're working with alpha fold!!)! 🥳 How did you learn bioinformatics? Are there any resources you'd recommend for a complete beginner? (I'm trying to learn how to do proteomics analyses rn, but I can't seem to find many resources I can easily understand 🙈)
hii!! thank you so much for the wishes! the paper was a part of my final project with my team + guide so it was really a group effort <3 my teammates and I are all first authors, they were just kind enough to put my name first xD and yes! my current alphafold work is a lot of fun ^=^ I'm really enjoying my internship hehe
okay so I learnt bioinformatics as a part of my uni curriculum, and we mainly relied on these two textbooks
Essential Bioinformatics by Jin Xiong
Bioinformatics: Sequence and Genome Analysis
In addition to this, I also recently found this textbook titled Bioinformatics: An Introductory Textbook by Thomas Dandekar, Meik Kunz which focuses both on basic bioinformatics concepts and how to use the available tools. I especially like this book because I found it super easy to understand
EMBL also offers courses! even for synchronous online/in person courses that have ended, they tend to upload course materials. There's one for proteomics analyses as well- I've checked out the mass spectrometry videos under this course and they start with the very basics
In addition to these, here are some playlists on youtube that've helped:
Foundations of Systems and Computational Biology, MIT OpenCourseWare // classroom videos, I'd suggest to check specific videos for a topic of your choice rather than actually going through all of them
Computational Systems Biology, IITM // videos on the basics of systems biology, networks and modelling
Bioinformatics 101 // videos on how to use a lot of tools, however this primarily focuses on gene expression and RNA seq data
lastly, I don't know if this would be particularly helpful given what you're looking for, but this textbook on Computational Genomics with R is a blessing for Omics related Data Analysis. So if you ever need to use R in a biological context, this would be a great place to start
yeah! so these are the majority of the resources that I've used, I hope they help! Just a heads up that the links to the textbooks I mentioned in the beginning are direct download links and won't take you to any website.
If there's anything else I can help with, feel free to reach out ^=^ best of luck!
#if anyone has come across any helpful resources for bioinformatics feel free to add to this post...#so much of academia is hidden behind a paywall it's really a shame#I hope these will be of some help!#i don't use coursera/edx/etc so I didn't add any of those links on here#studyblr#resources#answered#bioinformatics#bio student#biology#regarding cpb
30 notes
·
View notes
Text
NSH creating hunter

Explanation: poliphen-2 (in the drawing poliphen 7) is a bioinformatics tool used to predict how dangerous would a mutation be. It uses a scale from 0 to 1, being 1 the highest risk.
Edit: this has been in my drafts for like a year and idk why im made it anymore but i guess ill post it now?
#I don’t think sig gave hunter hyper cancer on purpose but also I don’t think a supercomputer could not detect it#also is funnier this way#rain world#no significant harassment#rw nsh#rw hunter#blue doodles
7 notes
·
View notes
Text
the real Scientist Experience is googling a bioinformatics tool and getting results for ice fishing and ice fishing only
6 notes
·
View notes
Text
I swear a solid 90% of bioinformatics is spent troubleshooting the bioinformatic tools
4 notes
·
View notes